Legend
Node information
Information about the selected node.
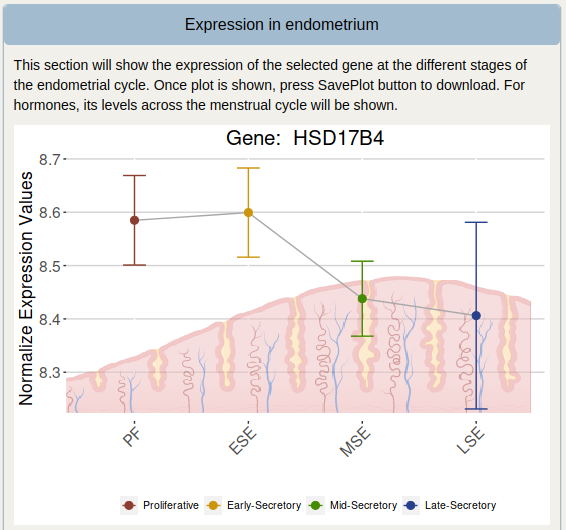
Expression in endometrium
This section shows the expression of the selected gene through the phases of the endometrial cycle. Once the plot is shown, press the SavePlot button to download the plot. For hormones, their levels across the menstrual cycle are shown.
Interactions
Information about the selected interactions.

To demonstrate the useful features of PROGESNET, the HSD17B4 gene is used as an example.
Nodes input
Gene or genes separated by commas (e.g., gene1,gene2) are typed into the Nodes text box using one of these gene ID formats:
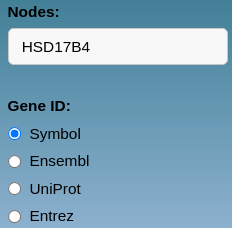
- Symbol: genes should be written either all in uppercase or all in lowercase (i.e., HSD17B4 or hsd17b4).
- Ensembl: "ENSG" or "ensg" should be written before the number code (i.e., ENSG00000133835 or ensg00000133835).
- UniProt: use the UniProt code in uppercase or lowercase (i.e., P51659 or p51659).
- Entrez: use the NCBI Entrez code only as numbers (i.e., 3295).
Hormones can also be added to the network by selecting one or more from the drop-down list.
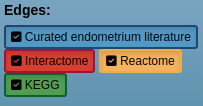
Edges from different sources can be filtered prior to the network search.
Click
 to show the network graph.
to show the network graph.
Network Graph
After clicking Run, the HSD17B4 network will be displayed. Green dots represent hormones, red squares represent genes whose expression has been measured in the endometrium in the phases of the menstrual cycle, blue squares represent genes expressed in endometrium and that have been experimentally validated using a single-molecule approach (Marti-Garcia et al., 2025), and grey squares represent genes with no information about their expression in endometrium. Edge colours are determined by the interaction source: blue edges represent data from the curated healthy endometrium literature (Marti-Garcia et al., 2025), red edges represent data from the human interactome, yellow edges represent data from the Reactome database and green edges represent data from the KEGG database.
The network edges can be filtered dynamically by selecting and deselecting the Edges sources from the left input panel.
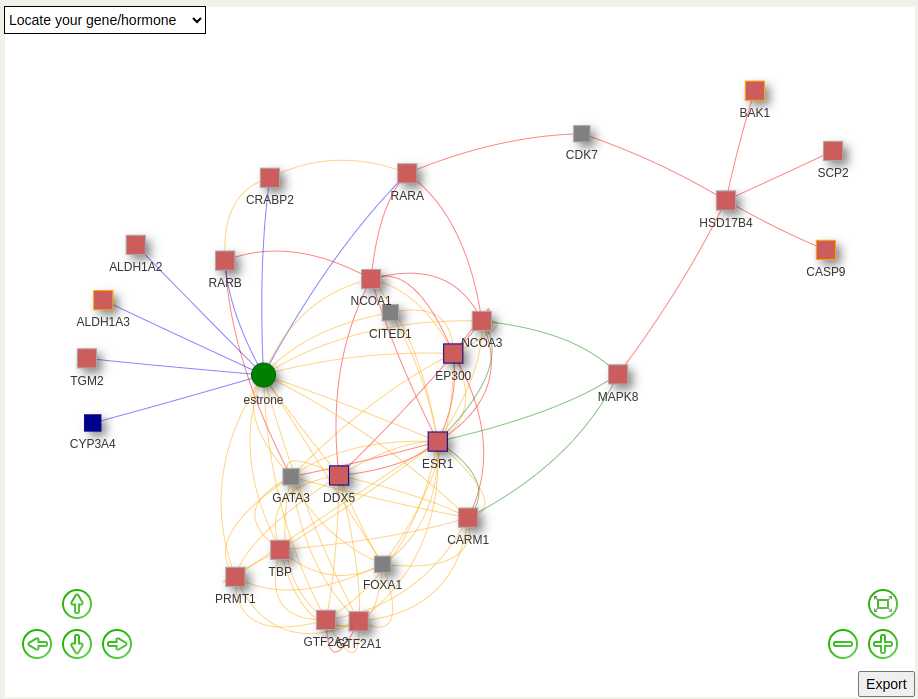
The network graph can be moved up and down and left and right using the keyboard arrows and can be zoomed in and out using the +/- keys.
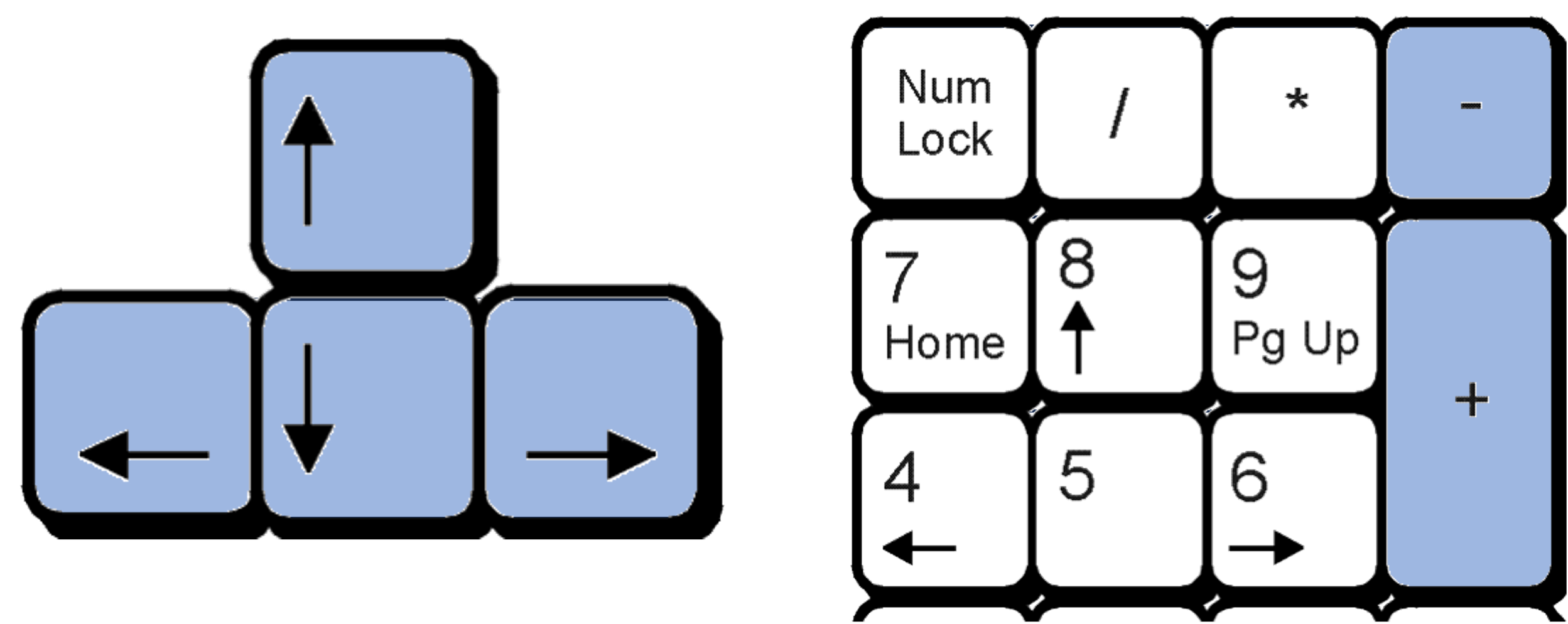


The network graph can be saved by clicking the Export button.
Interactions
All the interaction details of a selected node are displayed in this interactive table. Filters can be applied at the top of each column by typing in the corresponding text_box. The information for a single interaction can also be displayed by selecting it from the network.
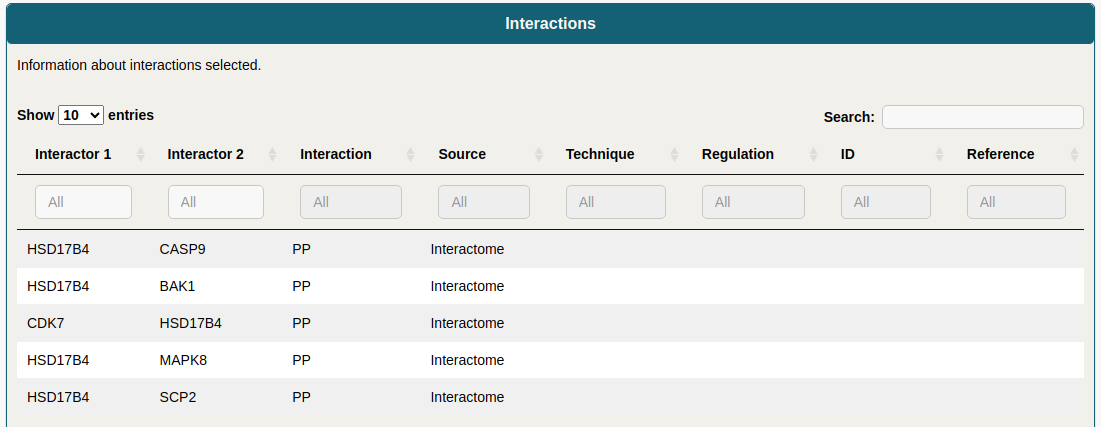
Some columns can be hidden by selecting them using the Column visibility button. The final table can be copied, printed or exported as .csv or .pdf files.
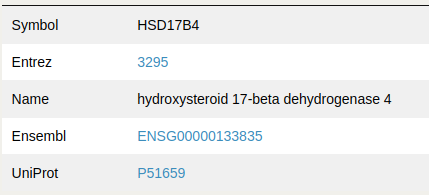
Nodes information
By clicking a node within the network, different gene nomenclatures are shown, including direct links to their corresponding official websites.
The expression throughout the endometrial cycle of the selected gene (if applicable) is displayed below the table. This plot can be downloaded by clicking the SavePlot button.
Frequently Asked Questions
Main publications of the group
-
Stratifying IVF population endometria using a prognosis gradient independent of endometrial timing.
Sanchez-Reyes JM, Parraga-Leo A, Sebastian-Leon P, Vidal MC, Marti-Garcia D, Spath K, Sanchez-Ribas I, Sanz FJ, Pellicer N, Remohi J, Wells D, Pellicer A, Diaz-Gimeno P. Hum Reprod. 2025 Aug 12;deaf156. doi: 10.1093/humrep/deaf156. -
Are we considering all the potential drug–drug interactions in women’s reproductive health? A predictive model approach.
Garcia-Acero P, Henarejos-Castillo I, Sanz FJ, Sebastian-Leon P, Parraga-Leo A, Garcia-Velasco JA, Diaz-Gimeno P. Pharmaceutics. 2025; 17(8):1020. doi: 10.3390/pharmaceutics17081020. -
Advanced maternal age was associated with an annual decline in reproductive success despite use of donor oocytes: a retrospective study.
Sebastian-Leon P, Sanz FJ, Molinaro P, Pellicer A, Diaz-Gimeno P. Fertil Steril. 2025 May 20. doi: 10.1016/j.fertnstert.2025.05.150. -
Whole-exome sequencing and Drosophila modelling reveal mutated genes and pathways contributing to human ovarian failure.
Henarejos-Castillo I, Sanz FJ, Solana-Manrique C, Sebastian-Leon P, Medina I, Remohi J, Paricio N, Diaz-Gimeno P. Reprod Biol Endocrinol. 2024 Dec 4;22(1):153. doi: 10.1186/s12958-024-01325-4. -
Age-related uterine changes and its association with poor reproductive outcomes: a systematic review and meta-analysis.
Marti-Garcia D, Martinez-Martinez A, Sanz FJ, Devesa-Peiro A, Sebastian-Leon P, Del Aguila N, Pellicer A, Diaz-Gimeno P. Reprod Biol Endocrinol. 2024 Nov 30;22(1):152. doi: 10.1186/s12958-024-01323-6. -
Corticosteroids, androgens, progestogens and estrogens in the endometrial microenvironment and their association with endometrial progression and function.
Marti-Garcia D, Devesa-Peiro A, Labarta E, Lopez Nogueroles M, Sebastian-Leon P, Pellicer N, Meseguer M, Diaz-Gimeno P. Reprod Biomed Online. 2024 Jul 15;49(6):104377. doi: 10.1016/j.rbmo.2024.104377. -
Deciphering a shared transcriptomic regulation and the relative contribution of each regulator type through endometrial gene expression signatures.
Parraga-Leo A, Sebastian-Leon P, Devesa-Peiro A, Marti-Garcia D, Pellicer N, Remohi J, Dominguez F, Diaz-Gimeno P. Reprod Biol Endocrinol. 2023 Sep 12;21(1):84. doi: .10.1186/s12958-023-01131-4. -
Predicted COVID-19 molecular effects on endometrium reveal key dysregulated genes and functions.
Henarejos-Castillo I, Devesa-Peiro A, de Miguel-Gomez L, Sebastian-Leon P, Romeu M, Aleman A, Molina-Gil C, Pellicer A, Cervello I, Diaz-Gimeno P. Mol Hum Reprod. 2022 Oct 7;28(11):gaac035. doi: 10.1093/molehr/gaac035. -
Use of the endometrial receptivity array to guide personalized embryo transfer after a failed transfer attempt was associated with a lower cumulative and per transfer live birth rate during donor and autologous cycles.
Cozzolino M, Diaz-Gimeno P, Pellicer A, Garrido N. Fertil Steril. 2022;118(4):724-736. doi: 10.1016/j.fertnstert.2022.07.007. -
Breaking the ageing paradigm in endometrium: endometrial gene expression related to cilia and ageing hallmarks in women over 35 years.
Devesa-Peiro A, Sebastian-Leon P, Parraga-Leo A, Pellicer A, Diaz-Gimeno P. Hum Reprod. 2022 Apr 1;37(4):762-776. doi: 10.1093/humrep/deac010. -
Identifying and optimizing human endometrial gene expression signatures for endometrial dating.
Diaz-Gimeno P, Sebastian-Leon P, Sanchez-Reyes JM, Spath K, Aleman A, Vidal C, Devesa-Peiro A, Labarta E, Sánchez-Ribas I, Ferrando M, Kohls G, García-Velasco JA, Seli E, Wells D, Pellicer A. Hum Reprod. 2022 Jan 28;37(2):284-296. doi: 10.1093/humrep/deab262.











